Implicit membrane modeling allows facile determination of the binding orientation of a peripheral membrane protein to a membrane. Typically, we take the crystal structure and place it on the membrane surface at 6 different orientations. We then run a few ns of MD simulations. In some cases we observe that the protein always reorients to adopt the same final orientation. In other cases, the final orientation from the 6 simulations can be different, in which case we can compare their effective energies to determine which one is optimal. This approach has been applied to a Fatty Acid Binding Protein (Ref. 1) and to a membrane-bound proteinase (Ref. 3).
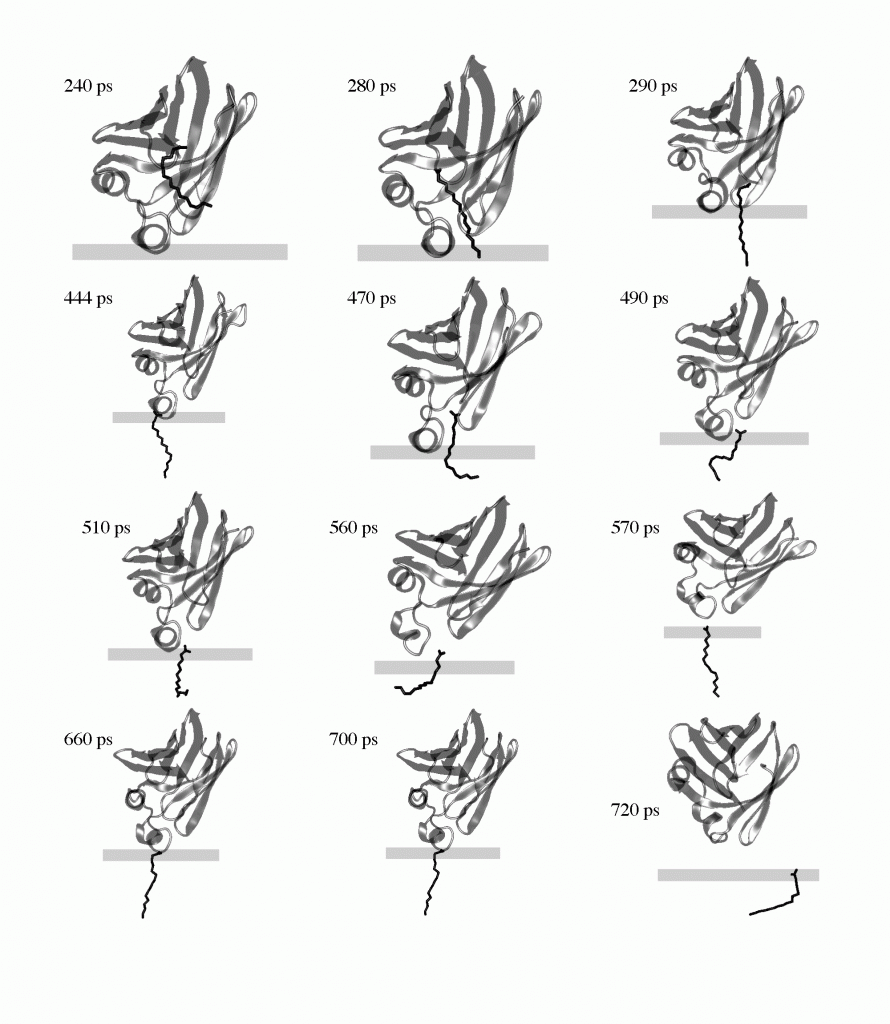
The transfer of palmitate from the membrane bound holo-IFABP to the membrane via the helical portal region. The membrane is represented by the rectangle; the upper surface corresponds to the plane of smeared charge; the lower surface is the hydrocarbon/headgroup boundary.
A similar approach was used to determine the membrane-bound structure of a-synuclein, a membrane that aggregates in Parkinson’s disease (Ref. 2). The protein was built as an ideal, straight a-helix and remained so during the simulations, although it exhibited considerable flexibility near the middle. The periodicity of the helix was analyzed and was found to be 11/3, instead of the 18/5 typically observed in aqueous helices.
References
- Mihajlovic, M., Lazaridis, T. “Modeling fatty acid delivery from intestinal fatty acid binding protein to a membrane”, Prot. Sci. , 16:2042-55 (2007)
- Mihajlovic, M., Lazaridis, T. “Membrane-bound structure and energetics of alpha-synuclein”, Proteins , 70:761-78 (2008)
- Hajjar, E., Mihajlovic, M., Witko-Sarsat, V., Lazaridis, T., Reuter “Computational prediction of the binding site of proteinase 3 to the plasma membrane”, Proteins , 71:1655-69 (2008)
